- Biophysics PhD candidate at Johns Hopkins.
- Advisor: Dr. Albert Lau
- Expected graduation: Summer 2024
My PhD research has centered around a key question: Given a protein structure, how can we computationally find ways of inhibiting the protein? Answering this question requires software development, protein simulation, and data analysis.
Research Projects
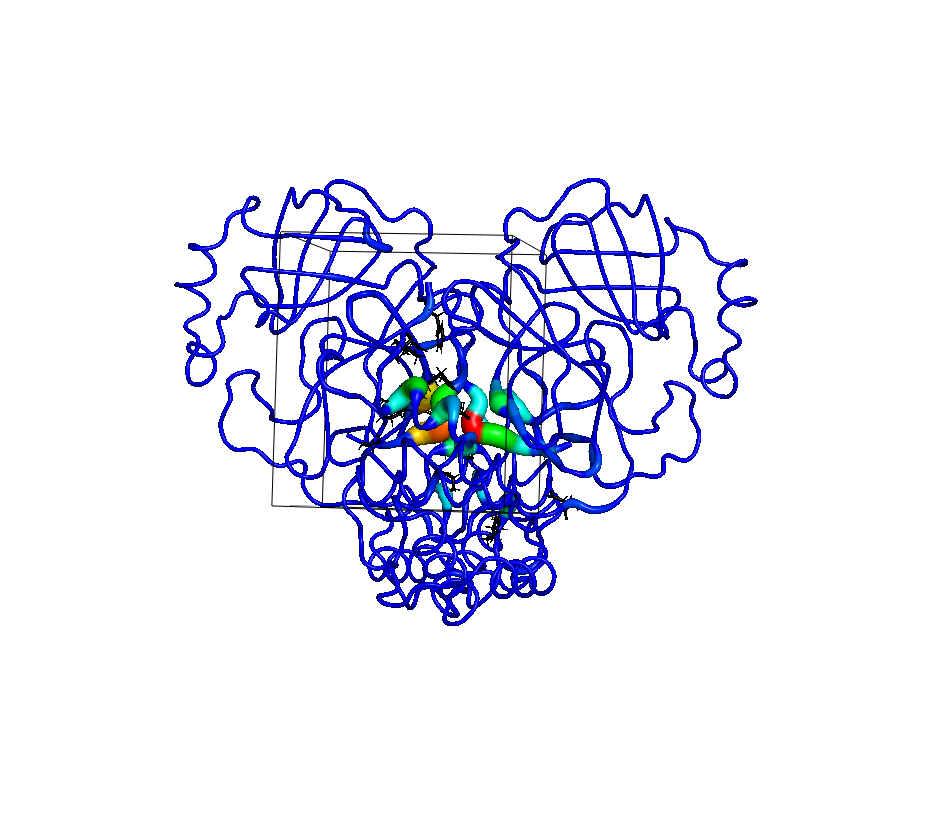
Binding site identification: I developed the TACTICS software to find binding sites within molecular dynamics simulations. TACTICS outperforms popular alternatives in speed and accuracy.
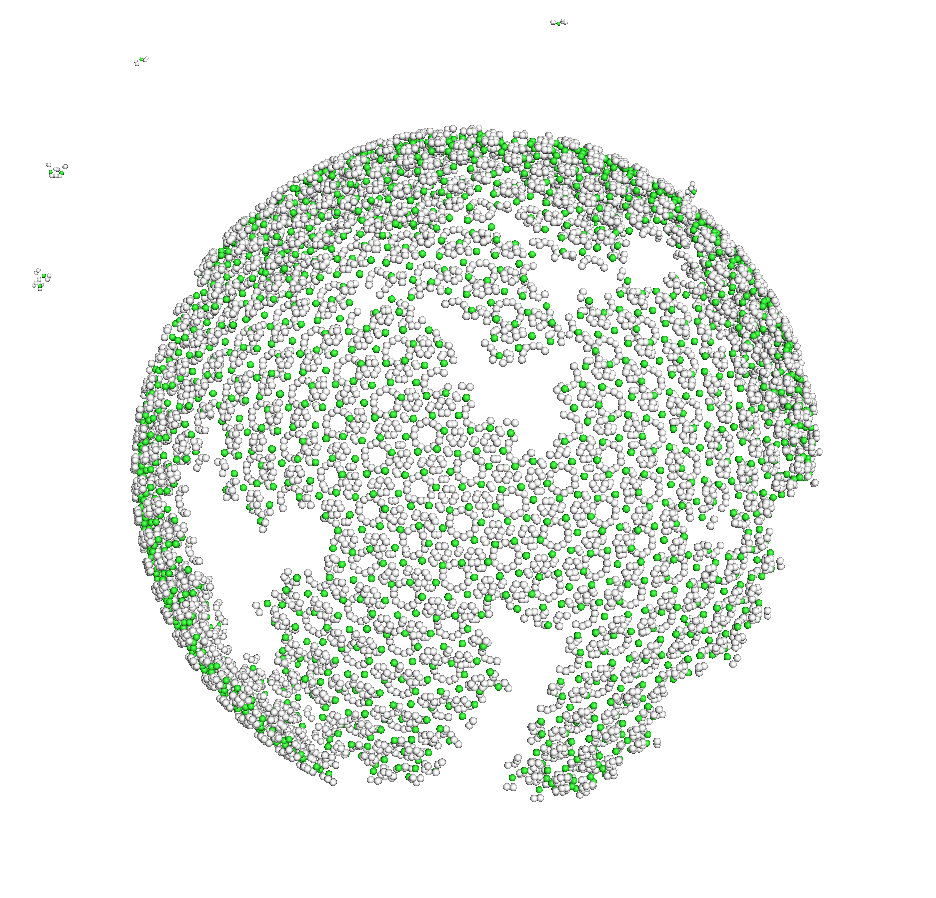
HIV gag self-assembly: I used a reaction-diffusion model to assess conditions required for HIV gag lattice assembly. The results suggest that HIV reproduction would be inhibited when Gag self-assembly conditions are shifted outside of a narrow range.

Allosteric signal transmission: I developed the EVADE software to quantify dihedral correlations. I used EVADE to propose an allosteric mechanism in the SARS-CoV-2 main protease. I also used EVADE to demonstrate common pitfalls in dihedral correlation analysis.
© 2024 Daniel James Evans. All rights reserved.